For the love of God – learn to touch-type
 I was in a moderately-well-known-Professor-who-shall-remain-nameless’s office the other day, watching him bang out an email. Except he wasn’t banging it out, he was using his two index fingers to hunt-and-peck at the keyboard while continually lifting his head to look at the screen, and then putting it back down to peer at the keyboard. It was painful to watch. I nearly gnawed right through a knuckle.
I was in a moderately-well-known-Professor-who-shall-remain-nameless’s office the other day, watching him bang out an email. Except he wasn’t banging it out, he was using his two index fingers to hunt-and-peck at the keyboard while continually lifting his head to look at the screen, and then putting it back down to peer at the keyboard. It was painful to watch. I nearly gnawed right through a knuckle.
I was fortunate. When I was 13 years old, my Mum sat me down at her electric typewriter (yes, really, I am that old), gave me a Mavis Beacon book, and told me I wasn’t allowed any dinner until I could do at least 60 words per minute with no mistakes. A week later, when I was so faint with hunger and the pain from my finger-blisters that I could barely see the page anymore, I managed it.
Of course that’s not true, but my Mum is a fantastic typist, and did teach me when I was about 13, and honestly in terms of investment/payoff ratio it’s probably the best few hours I’ve ever spent in my life. Learning to touch-type is not hard and doesn’t really take that long; like most things it’s just a matter of discipline and practice. Actually, once you start doing it properly it’s hard to imagine how you ever managed without it. If you’re reading this, then you probably spend at least a substantial part of your day sitting at a keyboard, why not spend a few hours making the entire rest of your working life easier and more efficient? And if you’re reading this thinking “Yeah, but I’ve developed my own version of semi-touch-typing which is pretty fast and efficient, actually” then you’re wrong. It’s likely nowhere near as fast and efficient as it could be.
There are lots of good online touch-typing courses available. Most of them give you feedback on speed and the number of mistakes you make while typing. TypingClub looks like a good option. The BBC has a good course aimed more at kids. This site has a free course and a number of games to improve and sharpen up your skillz.
Seriously, this is one of the best things you can possibly learn. Yes, it’s a drag, but it will literally make the rest of your life easier. DO IT.
Stimulus timing accuracy in PsychoPy – an update, and an example of open science in action
A few days ago I reported on a new paper that tested the timing accuracy of experimental software packages. The paper suggested that PsychoPy had some issues with presenting very fast stimuli that only lasted one (or a few) screen refreshes.
However, the creator of PsychoPy (Jon Peirce, of Nottingham University) has already responded to the data in the paper. The authors of the paper made all the data and (importantly) the program scripts used to generate it available on the Open Science Framework site. As a result it was possible for Jon to examine the scripts and see what the issue was. It turns out the authors used an older version of PsychoPy, where the ability to set a stimulus duration in frames (or screen refreshes) wasn’t available. Up-to-date versions have this feature, and as a result are now much more reliable. Those who need the full story should read Jon’s comment (and the response by the authors) on the PLoS comments page for the article.
So, great news if, like me, you’re a fan of PsychoPy. However, there’s a wider picture that’s revealed by this little episode, and a very interesting one I think. As a result of the authors posting their code up to the OSF website, and the fact that PLoS One allows comments to be posted on its articles, the issue could be readily identified and clarified, in a completely open and public manner, and within a matter of days. This is one of the best examples I’ve seen of the power of an open approach to science, and the ability of post-publication review to have a real impact.
Interesting times, fo’ shiz.
A new paper on timing errors in experimental software
 A new paper just out in PLoS One (thanks to Neuroskeptic for pointing it out on Twitter) shows the results of some tests conducted on three common pieces of software used in psychology and neuroscience research: DMDX, E-Prime, and PsychoPy. The paper is open-access, and you can read it here. The aim was to test the timing accuracy of the software when presenting simple visual stimuli (alternating black and white screens). As I’ve written about before, accurate timing in experiments can often be of great importance and is by no means guaranteed, so it’s good to see some objective work on this, conducted using modern hardware and software.
A new paper just out in PLoS One (thanks to Neuroskeptic for pointing it out on Twitter) shows the results of some tests conducted on three common pieces of software used in psychology and neuroscience research: DMDX, E-Prime, and PsychoPy. The paper is open-access, and you can read it here. The aim was to test the timing accuracy of the software when presenting simple visual stimuli (alternating black and white screens). As I’ve written about before, accurate timing in experiments can often be of great importance and is by no means guaranteed, so it’s good to see some objective work on this, conducted using modern hardware and software.
The authors followed a pretty standard procedure for this kind of thing, which is to use a piece of external recording equipment (in this case the Black Box Tool Kit) connected to a photo-diode. The photodiode is placed on the screen of the computer being tested and detects every black-to-white or white-to-black transition, and the data is logged by the BBTK device. This provides objective information about when exactly the visual stimulus was displayed on the screen (as opposed to when the software running the display thinks it was displayed on the screen, which is not necessarily the same thing). In this paper the authors tested this flickering black/white stimulus at various speeds from 1000ms, down to 16.6667ms (one screen refresh or ‘tick’ on a standard 60Hz monitor).
It’s a nice little paper and I would urge anyone interested to read it in full; the introduction has a really nice review of some of the issues involved, particularly about different display hardware. At first glance the results are a little disappointing though, particularly if (like me) you’re a fan of PsychoPy. All three bits of software were highly accurate at the slower black/white switching times, but as the stimulus got faster (and was therefore more demanding on the hardware) errors began to creep in. At times less than 100ms, errors (in the form of dropped, or added frames) begin to creep in to E-Prime and PsychoPy; DMDX on the other hand just keeps on truckin’, and is highly accurate even at the fastest-switch conditions. PsychoPy is particularly poor under the most demanding conditions, with only about a third of ‘trials’ being presented ‘correctly’ under the fastest condition.
Why does this happen? The authors suggest that DMDX is so accurate because it uses Microsoft’s DirectX graphics libraries, which are highly optimised for accurate performance on Windows. Likewise, E-Prime uses other features of Windows to optimise its timing. PsychoPy on the other hand is platform-agnostic (it will run natively on Windows, Mac OS X, and various flavours of Linux) and therefore uses a fairly high-level language (Python). In simple terms, PsychoPy can’t get quite as close to the hardware as the others, because it’s designed to work on any operating system; there are more layers of software abstraction between a PsychoPy script and the hardware.
Is this a problem? Yes, and no. Because of the way that the PsychoPy ‘coder’ interface is designed, advanced users who require highly accurate timing have the opportunity to optimise their code, based on the hardware that they happen to be using. There’s no reason why a Python script couldn’t take advantage of the timing features in Windows that make E-Prime accurate too – they’re just not included in PsychoPy by default, because it’s designed to work on unix-based systems too. For most applications, dropping/adding a couple of frames in a 100ms stimulus presentation is nothing in particular to worry about anyway; certainly not for the applications I mostly use it for (e.g. fMRI experiments where, of course, timing is important in many ways, but the variability in the haemodynamic response function tends to render a lot of experimental precision somewhat moot). The authors of this paper agree, and conclude that all three systems are suitable for the majority of experimental paradigms used in cognitive research. For me, the benefits of PsychoPy (cross-platform, free licensing, user-friendly interface) far outweigh the (potential) compromise in accuracy. I haven’t noticed any timing issues with PsychoPy under general usage, but I’ve never had a need to push it as hard as these authors did for their testing purposes. It’s worth noting that all the testing in this paper was done using a single hardware platform; possibly other hardware would give very different results.
Those who are into doing very accurate experiments with very short display times (e.g. research on sub-conscious priming, or visual psychophysics) tend to use pretty specialised and highly-optimised hardware and software, anyway. If I ever had a need for such accuracy, I’d definitely undertake some extensive testing of the kind that these authors performed, no matter what hardware/software I ended up using. As always, the really important thing is to be aware of the potential issues with your experimental set-up, and do the required testing before collecting your data. Never take anything for granted; careful testing, piloting, examination of log files etc. is a potential life-saver.
TTFN.
Towards open-source psychology research
 A couple of interesting things have come along recently which have got me thinking about the ways in which research is conducted, and how software is used in psychology research.
A couple of interesting things have come along recently which have got me thinking about the ways in which research is conducted, and how software is used in psychology research.
The first is some recent publicity around the Many Labs replication project – a fantastic effort to try and perform replications of some key psychological effects, with large samples, and in labs spread around the world. Ed Yong has written a really great piece on it here for those who are interested. The Many Labs project is part of the Open Science Framework – a free service for archiving and sharing research materials (data, experimental designs, papers, whatever).
The second was a recent paper by Tom Stafford and Mike Dewar in Psychological Science. This is a really impressive piece of research from a very large sample of participants (854,064!) who played an online game. Data from the game was analysed to provide metrics of perception, attention and motor skills, and to see how these improved with training (i.e. more time spent playing the game). The original paper is here (paywalled, unfortunately), but Tom has also written about it on the Mind Hacks site and on his academic blog. The latter piece is interesting (for me anyway) as Tom says that he found his normal approach to analysis just wouldn’t work with this large a dataset and he was obliged to learn Python in order to analyse the data. Python FTW!
Anyway, the other really nice thing about this piece of work is that the authors have made all the data, and the code used to analyse it, publicly available in a GitHub repository here. This is a great thing to do, particularly for a large, probably very rich dataset like this – potentially there are a lot of other analyses that could be run on these data, and making it available enables other researchers to use it.
These two things crystallised an important realisation for me: It’s now possible, and even I would argue preferential, for the majority of not-particularly-technically-minded psychology researchers to perform their research in a completely open manner. Solid, free, user-friendly cross-platform software now exists to facilitate pretty much every stage of the research process, from conception to analysis.
Some examples: PsychoPy is (in my opinion) one of the best pieces of experiment-building software around at the moment, and it’s completely free, cross-platform, and open-source. The R language for statistical computing is getting to be extremely popular, and is likewise free, cross-platform, etc. For analysis of neuroimaging studies, there are several open-source options, including FSL and NiPype. It’s not hard to envision a scenario where researchers who use these kinds of tools could upload all their experimental files (experimental stimulus programs, resulting data files, and analysis code) to GitHub or a similar service. This would enable anyone else in the world who had suitable (now utterly ubiquitous) hardware to perform a near-as-dammit exact replication of the experiment, or (more likely) tweak the experiment in an interesting way (with minimal effort) in order to run their own version. This could potentially really help accelerate the pace of research, and the issue of poorly-described and ambiguous methods in papers would become a thing of the past, as anyone who was interested could simply download and demo the experiment themselves in order to understand what was done. There are some issues with uploading very large datasets (e.g. fMRI or MEG data) but initiatives are springing up, and the problem seems like it should be a very tractable one.
The benefit for researchers should hopefully be greater visibility and awareness of their work (indexed in whatever manner; citations, downloads, page-views etc.). Clearly some researchers (like the authors of the above-mentioned paper) have taken the initiative and are already doing this kind of thing. They should be applauded for taking the lead, but they’ll likely remain a minority unless researchers can be persuaded that this is a good idea. One obvious prod would be if journals started encouraging this kind of open sharing of data and code in order to accept papers for publication.
One of the general tenets of the open-source movement (that open software benefits everyone, including the developers) is doubly true of open science. I look forward to a time when the majority of research code, data, and results are made public in this way and the research community as a whole can benefit from it.
How to hack conditional branching in the PsychoPy builder
Regular readers will know that I’m a big fan of PsychoPy, which (for non-regular readers; *tsk*) is a piece of free, open-source software for designing and programming experiments, built on the Python language. I’ve been using it a lot recently, and I’m happy to report my initial ardour for it is still lambently undimmed.
The PsychoPy ‘builder’ interface (a generally brilliant, friendly, GUI front-end) does have one pretty substantial drawback though; it doesn’t support conditional branching. In programming logic, a ‘branch’ is a point in a program which causes the computer to start executing a different set of instructions. A ‘conditional branch’ is where the computer decides what to do out of two of more alternatives (i.e. which branch to follow) based on some value or ‘condition’. Essentially the program says, ‘if A is true: do X, otherwise (or ‘else’ in programming jargon) if B is true; do Y’. One common use of conditional branching in psychology experiments is to repeat trials that the subject got incorrect; for instance, one might want one’s subjects to achieve 90% correct on a block of trials before they continue to the next one, so the program would have something in it which said ‘if (correct trials > 90%); then continue to the next block, else if (correct trials < 90%); repeat the incorrect trials’.
At the bottom of the PsychoPy builder is a time-line graphic (the ‘flow panel’) which shows the parts of the experiment:
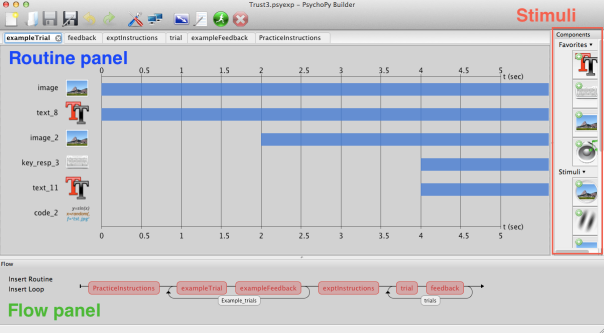 The experiment proceeds from left to right, and each part of the flow panel is executed in turn. The loops around parts of the flow panel indicate that the bits inside the loops are run multiple times (i.e. they’re the trial blocks). This is an extremely powerful interface, but there’s no option to ‘skip’ part of the flow diagram – everything is run in the order in which it appears from left-to-right.
The experiment proceeds from left to right, and each part of the flow panel is executed in turn. The loops around parts of the flow panel indicate that the bits inside the loops are run multiple times (i.e. they’re the trial blocks). This is an extremely powerful interface, but there’s no option to ‘skip’ part of the flow diagram – everything is run in the order in which it appears from left-to-right.
This is a slight issue for programming fMRI experiments that use block-designs. In block-design experiments, typically two (or more) blocks of about 15-20 seconds are alternated. They might be a ‘rest’ block (no stimuli) alternated with a visual stimulus, or two different kinds of stimuli, say, household objects vs. faces. For a simple two-condition-alternating experiment one could just produce routines for two conditions, and throw a loop around them for as many repeats as needed. The problem arises when there are more than two block-types in your experiment, and you want to randomise them (i.e. have a sequence which goes ABCBCACAB…etc.). There’s no easy way of doing this in the builder. In an experiment lasting 10 minutes one might have 40 15-second blocks, and the only way to produce the (psuedo-random) sequence you want is with 40 separate elements in the flow panel that all executed one-by-one (with no loops). Building such a task would be very tedious, and more importantly, crashingly inelegant. Furthermore, you probably wouldn’t want to use the same sequence for every participant, so you’d have to laboriously build different versions with different sequences of the blocks. There’s a good reason for why this kind of functionality hasn’t been implemented; it would make the builder interface much more complicated and the PsychoPy developers are (rightly) concerned with keeping the builder as clean and simple as possible.
Fortunately, there’s an easy little hack which was actually suggested by Jon Peirce (and others) on the PsychoPy users forum. You can in fact get PsychoPy to ‘skip’ routines in the flow panel, by the use of loops, and a tiny bit of coding magic. I thought it was worth elaborating the solution on here somewhat, and I even created a simple little demo program which you can download and peruse/modify.
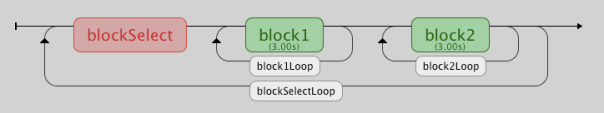
So, this is how it works. I’ve set up my flow panel like this:
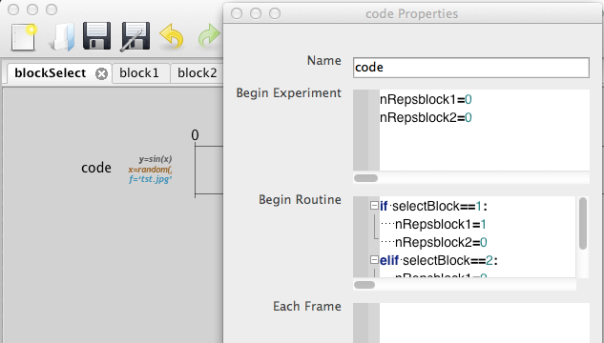
So, there are two blocks, each of which have their own loop, a ‘blockSelect’ routine, and a ‘blockSelectLoop’ enclosing the whole thing. The two blocks can contain any kind of (different) stimulus element; one could have pictures, and one could have sounds, for instance – I’ve just put some simple text in each one for demo purposes. The two block-level loops have no condition files associated with them, but in the ‘nReps’ field of their properties box I’ve put a variable ‘nRepsblock1’ for block1 and ‘nRepsblock2’ for block2. This tells the program how many times to go around that loop. The values of these variables are set by the blockSelect routine which contains a code element, which looks like this:
The full code in the ‘Begin Routine’ box above is this:
if selectBlock==1: nRepsblock1=1 nRepsblock2=0 elif selectBlock==2: nRepsblock1=0 nRepsblock2=1
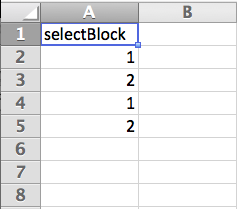
This is a conditional branching statement which says ‘if selectBlock=1, do X, else if selectBlock=2, do Y’. The variable ‘selectBlock’ is derived from the conditions file (an excel workbook) for the blockSelectLoop, which is very simple and looks like this:
So, at the beginning of the experiment I define the two variables for the number of repetitions for the two blocks, then on every go around the big blockSelectLoop, the code in the blockSelect routine sets one of the number of repetitions of each of the small block-level loops to 0, and the other to 1. Setting the number of repetitions for a loop to 0 basically means ‘skip that loop’, so one is always skipped, and the other one is always executed. The blockSelectLoop sequentially executes the conditions in the excel file, so the upshot is that this program runs block1, then block2, then block1 again, then block2 again. Now all I have to do if I want to create a different sequence of blocks is to edit the column in the conditions excel file, to produce any kind of sequence I want.
Hopefully it should be clear how to extend this very simple example to use three (or more) block/trial types. I’ve actually used this technique to program a rapid event-related experiment based on this paper, that includes about 10 different trial types, randomly presented, and it works well. I also hope that this little program is a good example of what can be achieved by using code-snippets in the builder interface; this is a tremendously powerful feature, and really extends the capabilities of the builder well beyond what’s achievable through the GUI. It’s a really good halfway step between relying completely on the builder GUI and the scaryness of working with ‘raw’ python code in the coder interface too.
If you want to download this code, run it yourself and poke it with a stick a little bit, I’ve made it available to download as a zip file with everything you need here. Annoyingly, WordPress doesn’t allow the upload of zip files to its blogs, so I had to change the file extension to .pdf; just download (right-click the link and ‘Save link as…’) and then rename the .pdf bit to .zip and it should work fine. Of course, you’ll also need to have PsychoPy installed as well. Your mileage may vary, any disasters that occur as a result of you using this program are your own fault, etc. etc. blah blah.
Happy coding! TTFN.
3D printed response box. The future. It’s here.
This may not look like much, but it’s actually pretty cool. It’s a new five-button response-box being built for our MRI scanner by one of the technicians where I work. The cool thing is that the main chassis has been custom-designed, and then fabricated using a polyethylene-extruding 3D printer. The micro-switches for the four fingers and thumb and the wired connections for each have obviously been added afterwards.
3D printing in plastic is a great way of creating hardware for use in the MRI environment, as, well… it’s plastic, and it can create almost any kind of structure you can think of. We sometimes need to build custom bits of hardware for use in some of our experiments, and previously we’d usually build things by cutting plastic sheets or blocks to the required shape, and holding them together with plastic screws. Using a 3D-printer means we can produce solid objects which are much stronger and more robust, and they can be produced much more quickly and easily too. I love living in the future.
High-tech version of the hollow-face illusion
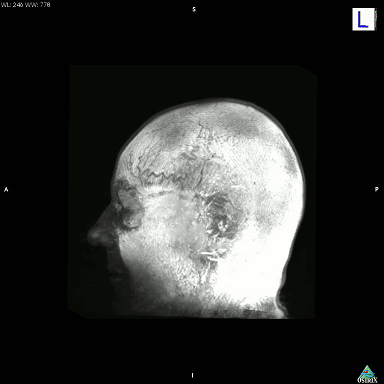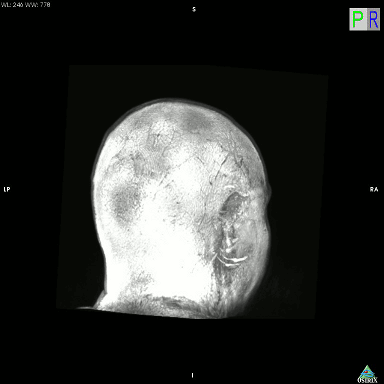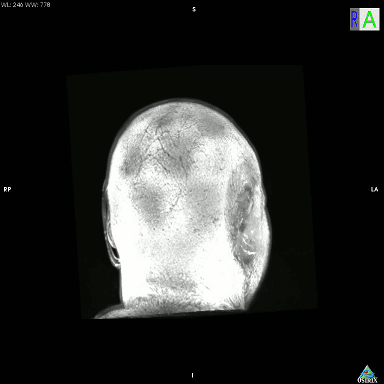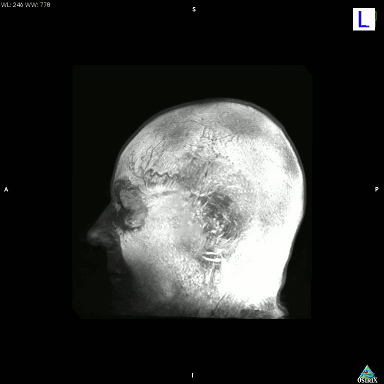
I just accidentally made a kind-of version of the classic hollow-face illusion, using an anatomical MRI scan of my own head and Osirix. I exported a movie in Osirix, saved the movie as an image sequence using Quicktime, and then assembled it into an animated GIF using GIMP.
This is a maximum intensity projection, and because of the way MIPs work, it appears that the head is rotating 180 degrees to the left, and then the direction switches and it rotates back 180 degrees to the right. In actual fact, the image is rotating constantly in one direction, as can be seen by looking at the cube on the top left which cycles through L (Left), P (Posterior), R (Right), and A (Anterior). It’s not really a hollow-face illusion as the effect is pretty much an artefact of the MIP, but still, I thought it was cool.
This page on how to design and make scientific conference posters…
…is really, really brilliant, and you should all go and look at it. That is all.
Some Python resources

Since I’ve switched to using PsychoPy for programming my behavioural and fMRI experiments (and if you spend time coding experiments, I strongly suggest you check it out too, it’s brilliant) I’ve been slowly getting up to speed with the Python programming language and syntax. Even though the PsychoPy GUI ‘Builder’ interface is very powerful and user-friendly, one inevitably needs to start learning to use a bit of code in order to get the best out of the system.
Before, when people occasionally asked me questions like “What programming language should I learn?” I used to give a somewhat vague answer, and say that it depended largely on what exactly they wanted to achieve. Nowadays, I’m happy to recommend that people learn Python, for practically any purpose. There are an incredible number of libraries available that enable you to do almost anything with it, and it’s flexible and powerful enough to fit a wide variety of use cases. Many people are now using it as a free alternative to Matlab, and even using it for ‘standard’ statistical analyses too. The syntax is incredibly straight-forward and sensible; even if an individual then goes on to use a different language, I think Python is a great place to start with programming for a novice. Python seems to have been rapidly adopted by scientists, and there are some terrific resources out there for learning Python in general, and its scientific applications in particular.
For those getting started there are a number of good introductory resources. This ‘Crash Course in Python for Scientists’ is a great and fairly brief introduction which starts from first principles and doesn’t assume any prior knowledge. ‘A Non-Programmers Tutorial for Python 2.6′ is similarly introductory, but covers a bit more material. ‘Learn Python the Hard Way’ is also a well-regarded introductory course which is free to view online, but has a paid option ($29.95) which gives you access to additional PDFs and video material. The ‘official’ Python documentation is also pretty useful, and very comprehensive, and starts off at a basic level. Yet another good option is Google’s Python Classes.
For those who prefer a more interactive experience, CodeAcademy has a fantastic set of interactive tutorials which guide you through from the complete beginning, up to fairly advanced topics. PythonMonk and TryPython.org also have similar systems, and all three are completely free to access – well worth checking out.
For Neuroimagers, there are some interesting Python tools out there, or currently under development. The NIPY (Neuroimaging in Python) community site is well worth a browse. Most interestingly (to me, anyway) is the nipype package, which is a tool that provides a standard interface and workflow for several fMRI analysis packages (FSL, SPM and FreeSurfer) and facilitates interaction between them – very cool. fMRI people might also be very interested in the PyMVPA project which has implemented various Multivariate Pattern Analysis algorithms.
People who want to do some 3D programming for game-like interfaces or experimental tasks will also want to check out VPython (“3D Programming for ordinary mortals”!).
Finally, those readers who are invested in the Apple ecosystem and own an iPhone/iPad will definitely want to check out Pythonista – a full featured development environment for iOS, with a lot of cool features, including exporting directly to XCode (thanks to @aechase for pointing this one out on Twitter). There looks to be a similar app called QPython for Android, though it’s probably not as full-featured; if you’re an Android user, you’re probably fairly used to dealing with that kind of disappointment though. ;o)
Anything I’ve missed? Let me know in the comments and I’ll update the post.
Toodles.

JASP might finally be the SPSS replacement we’ve been waiting for
Nov 27
Posted by Matt Wall
So, why do I keep using it? Because a) It’s what I learned as an undergraduate/PhD student and I know it backwards, and b) there are few viable alternatives. Yes, I know I should learn R, but I actually don’t use ‘normal’ stats that often (I spend most of my analysis time in brain-imaging packages these days) and every time I learn how to do something in R, I try doing it again a month later, have forgotten it, and have to learn it all over again. At some point I hope to become an R master, but for occasional use, I find the learning curve to be too steep. I would also hesitate to try and use R to teach students; I find it generally pretty user-hostile.
So, for ages now, I’ve been looking for a good, user-friendly, open-source alternative to SPSS. One that isn’t a bloated monster, but has enough features to enable basic analyses. I was quite hopeful about PSPP for a while (free software that tries to replicate SPSS as closely as possible). However it lacks some relatively basic ANOVA features, and since one of the things I dislike about SPSS is the interface, trying to replicate it seems like a bit of a mistake. SOFA statistics was a contender too, and it does have a beautiful interface and produce very nice-looking results, but it only does one-way ANOVAS, so… fail.
So, I gave up and crawled miserably back to SPSS. However, fresh hope now burns within my chest, as the other day I came across JASP (which the developers insist, definitely does not stand for ‘Just Another Statistics Program’). The aim of JASP is to be ‘a low fat alternative to SPSS, a delicious alternative to R.’ Nice. It seems to cover all the analysis essentials (t-test, ANOVA, regression, correlation) plus also has some fancier Bayesian alternatives and a basic Structural Equation Modelling option. The interface is great, and the results tables update in real-time as you change the options in your analysis! Very nice. This demo video gives a good overview of the features and workflow:
It’s clearly very much a work-in-progress. One issue is that it doesn’t have any in-built tools for data manipulation. It will read .csv text files, but they basically have to be in a totally ready-to-analyse format, which means general data-cleaning/munging procedures have to be done in Excel/Matlab/R/whatever. Another major downside is that there appears to be no facility for saving or scripting analysis pipelines. Hopefully though, development will continue and other features will gradually appear… I’ll be keeping a close eye on it!
Posted in Commentary, Software, Study Skills
12 Comments
Tags: analysis, JASP, PSPP, R, R stats, SPSS, statistical analysis, statistics